library(gamboostLSS)13: Variable Selection for Gene Expression Data
Introduction
This application uses the riboflavin data from the hdi package.
library(hdi)
data("riboflavin")
# number of observations and gene expression levels
dim(riboflavin$x)[1] 71 4088Coefficient paths
In Figure 13.1 of the book, we display the coefficient path for the first 100 iterations when fitting a Gaussian location and scale model.
dat <- riboflavin
# change the variable names
attr(dat$x, "dimnames")[[2]] <- paste("", 1:4088, sep = "")
# fit a boosting model (remove intercept for nicer plot)
glm1 <- glmboostLSS( y ~ - 1+ x, data = dat,
method = "noncyclic",
families = GaussianLSS(mu = NULL,
sigma = NULL,
stabilization = "MAD"),
control = boost_control(nu = 0.1))
par(mfrow = c(1,2))
plot(glm1, off2int = TRUE, col = 1, las = 1, ylim =c(-1,1))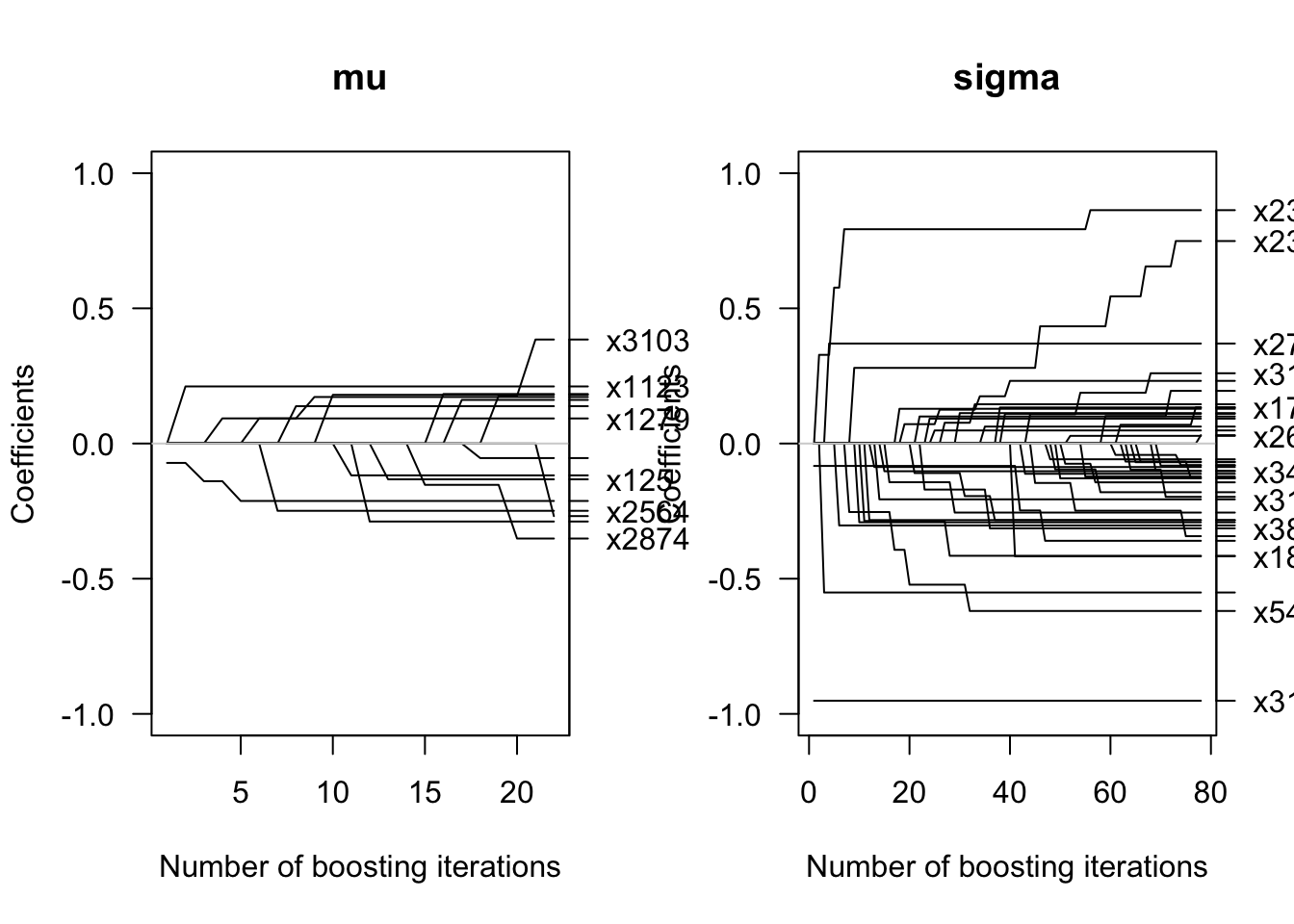
Leave-one-out cross-validation
We compare a classical boosting approach (mean regression) with a Gaussian location and scale and a Gumbel model. We use separate functions to compare the approaches running in parallel on a computing cluster.
First classical boosting
AMP <- function(id)
{
set.seed(id)
print(id)
# classic L2 boosting without one obs (id)
glm1 <- glmboost( y ~ x, data = dat[- id,])
# bootstrapping
cvr <- cvrisk(glm1, grid = 1:500, papply = "lapply")
mstop(glm1) <- mstop(cvr)
# predict on the left out obs
pred <- predict(glm1, newdata = dat[id,])
# compute msep
msep <- (dat[id,]$y - pred)^2
coefs <- coef(glm1, which = "")
return(list(pred = pred, msep = msep,
mstop = mstop(cvr), coef = coefs ))
}For executing in parallel
library(parallel)
id <- 1:nrow(dat)
ERG <- mclapply(id, FUN = AMP, mc.cores = 11,
mc.preschedule = FALSE)Now check the MSEP, MAEP and selcted variables:
mean(unlist(sapply(ERG, function(x)x[2]))) # msep[1] 0.2033454median(sqrt(unlist(sapply(ERG, function(x)x[2])))) # maep[1] 0.2405357mean(sapply(ERG, function(x) sum(x[4]$coef != 0)))[1] 33.21127Gaussian location and scale model
AMP_LSS <- function(id)
{
set.seed(id)
print(id)
glm1 <- glmboostLSS( y ~ x, data = dat[- id,],
method = "noncyclic",
families = GaussianLSS(),
control = boost_control(
mstop = 50))
cvr <- cvrisk(glm1, papply = lapply)
mstop(glm1) <- mstop(cvr)
pred <- predict(glm1, newdata = dat[id,],
type = "response")
msep <- (dat[id,]$y - pred$mu)^2
mae <- abs(dat[id,]$y - pred$mu)
coefs <- coef(glm1, which = "")
return(list(pred = pred, msep = msep,
mstop = mstop(glm1),
coef = coefs ))
}Execute in parallel
ERG <- mclapply(id, FUN = AMP_LSS, mc.cores = 11,
mc.preschedule = FALSE)mean(unlist(sapply(ERG, function(x)x[2]))) # msep[1] 0.2140843median(sqrt(unlist(sapply(ERG, function(x)x[2])))) # maep[1] 0.2342696mean(sapply(ERG, function(x) sum(x[4]$coef$mu != 0)))[1] 19.35211mean(sapply(ERG, function(x) sum(x[4]$coef$sigma != 0)))[1] 3.014085Gumbel distribution
AMP_LSS_GU <- function(id)
{
set.seed(id)
print(id)
glm1 <- glmboostLSS( y ~ x, data = dat[- id,],
method = "noncyclic",
families = as.families("GU",
stabilization = "MAD"),
control = boost_control(
mstop = 1, nu = 0.01))
cvr <- cvrisk(glm1, grid = 1:300, papply = lapply)
mstop(glm1) <- mstop(cvr)
pred <- predict(glm1, newdata = dat[id,],
type = "response")
msep <- (dat[id,]$y - pred$mu)^2
coefs <- coef(glm1, which = "")
return(list(pred = pred, msep = msep,
mstop = mstop(glm1), coef = coefs ))
}After running in parallel:
ERG <- mclapply(id, FUN = AMP_LSS, mc.cores = 11,
mc.preschedule = FALSE)mean(as.numeric(unlist(sapply(ERG, function(x)x[2]))))# msep[1] 0.3758644median(sqrt(as.numeric(unlist(sapply(ERG, function(x)x[2])))))# msep[1] 0.3587012mean(sapply(ERG, function(x) sum(x[4]$coef$mu != 0)))[1] 24.53521mean(sapply(ERG, function(x) sum(x[4]$coef$sigma != 0)))[1] 9.042254Comparison with cyclical approach
AMP_LSS_cyc <- function(id)
{
set.seed(id)
print(id)
glm1 <- glmboostLSS( y ~ x, data = dat[- id,],
method = "cyclic",
families =
GaussianLSS(
stabilization = "MAD"),
control = boost_control(
mstop = 200, nu = 0.01))
cvr <- cvrisk(glm1, papply = lapply)
mstop(glm1) <- mstop(cvr)
pred <- predict(glm1, newdata = dat[id,],
type = "response")
msep <- (dat[id,]$y - pred$mu)^2
mae <- abs(dat[id,]$y - pred$mu)
coefs <- coef(glm1, which = "")
return(list(pred = pred, msep = msep,
mstop = mstop(glm1),
coef = coefs ))
}Executing as above
ERG <- mclapply(id, FUN = AMP_LSS_cyc, mc.cores = 11,
mc.preschedule = FALSE)# removing one failing run
ERG[[43]] <- NULL
mean(as.numeric(unlist(sapply(ERG, function(x)x[2]))))# msep[1] 0.2449231median(sqrt(as.numeric(unlist(sapply(ERG, function(x)x[2])))))# msep[1] 0.2955776mean(sapply(ERG, function(x) sum(x[4]$coef$mu != 0)))[1] 28.18571mean(sapply(ERG, function(x) sum(x[4]$coef$sigma != 0)))[1] 6.742857One standard error rule
AMP_LSS_ose <- function(id)
{
set.seed(id)
print(id)
glm1 <- glmboostLSS( y ~ x, data = dat[- id,],
method = "noncyclic",
families = GaussianLSS(),
control =
boost_control(mstop = 1))
cvr <- cvrisk(glm1, grid = 1:100, papply = lapply)
means <- apply(cvr, mean, MAR = 2)
mstop_orig <- which.min(means)
risk_orig <- min(means)
ose <- sd(cvr[,mstop(cvr)])/sqrt(25)
mstop_ose <- min(which(means <= min(means) + ose))
mstop(glm1) <- mstop_ose
pred <- predict(glm1, newdata = dat[id,],
type = "response")
msep <- (dat[id,]$y - pred$mu)^2
mae <- abs(dat[id,]$y - pred$mu)
coefs <- coef(glm1, which = "")
selmu <- length(coef(glm1)$mu)-1
selsi <- length(coef(glm1)$sigma)-1
ls_ose<- dnorm(x = ribo[id,'y'],
mean = pred$mu,
sd = pred$sigma, log = TRUE)
return(list(pred = pred, msep = msep,
mstop = mstop(glm1),
selmu = selmu, selsi = selsi,
ls_ose = ls_ose))
}ERG <- mclapply(id, FUN = AMP_LSS_ose, mc.cores = 11,
mc.preschedule = FALSE)mean(unlist(sapply(ERG, function(x)x$selmu)))[1] 15mean(unlist(sapply(ERG, function(x)x$selsi)))[1] 1.43662mean(unlist(sapply(ERG, function(x)x$msep)))[1] 0.2848565median(sqrt(unlist(sapply(ERG, function(x)x$msep))))[1] 0.288833median(unlist(sapply(ERG, function(x)x$mstop)))[1] 36mean(unlist(sapply(ERG, function(x)x$ls_ose)))[1] -0.7925668Probing
AMP_LSS_probing <- function(id)
{
set.seed(id)
print(id)
glm1 <- glmboostLSS( y ~ x, data = dat[- id,],
method = "noncyclic",
families = GaussianLSS(),
control = boost_control(
mstop = 1))
set.seed(id)
probes.train <- as.data.frame(
sapply(ribo[,-1], sample)) # shuffling all variables (leaving out response which is at position 1 and 2)
names(probes.train) <- paste(names(ribo)[-c(1)],
"probe", sep = "_")
probes.train <- cbind(ribo, probes.train)
glm1 <- glmboostLSS( y ~ ., data = probes.train[- id,],
method = "noncyclic",
families = GaussianLSS(),
control = boost_control(
mstop = 100))
if(!any(selected(glm1,merge = T) >
length(names(ribo)[-c(1)]))) glm1[100]
mstop_probes <- min(which(selected(glm1,merge = T) >
length(names(ribo)[-c(1)]))) - 1
glm1[mstop_probes]
coefs <- coef(glm1)
selmu <- length(coef(glm1)$mu)-1
selsi <- length(coef(glm1)$sigma)-1
pred_mu <- predict(glm1$mu, newdata = probes.train[id,],
type = "response")
pred_sigma <- predict(glm1$sigma,
newdata = probes.train[id,],
type = "response")
msep_prob <- (dat[id,]$y - pred_mu)^2
mae_prob <- abs(dat[id,]$y - pred_mu)
ls_prob <- dnorm(x = ribo[id,'y'],
mean = pred_mu, sd = pred_sigma,
log = TRUE)
return(list(selmu = selmu, selsi = selsi,
mstop = mstop_probes,
pred_mu=pred_mu,
pred_sigma =pred_sigma,
msep = msep_prob, mae = mae_prob,
ls_prob = ls_prob))
}ERG <- mclapply(id, FUN = AMP_LSS_probing, mc.cores = 11,
mc.preschedule = FALSE)mean(unlist(sapply(ERG, function(x)x$selmu)))[1] 5.295775mean(unlist(sapply(ERG, function(x)x$selsi)))[1] 0.3661972mean(unlist(sapply(ERG, function(x)x$msep)))[1] 0.5452482median(sqrt(unlist(sapply(ERG, function(x)x$msep))))[1] 0.4737379median(unlist(sapply(ERG, function(x)x$mstop)))[1] 16mean(unlist(sapply(ERG, function(x)x$ls_prob)))[1] -1.136055Stability selection
AMP_LSS_stabsel <- function(id)
{
set.seed(id)
print(id)
glm1 <- glmboostLSS( y ~ x, data = dat[- id,],
method = "noncyclic",
families = GaussianLSS(),
control = boost_control(mstop = 100))
set.seed(id)
s1 <- stabsel(glm1, q = 20, PFER = 1, assumption ='none')
sel = names(s1$selected)
if(any(sel %in% (grep("(Intercept)",sel , value = T)))){
sel <- sel[-which(sel %in% (grep("(Intercept)", sel, value = T)))]
}
if(any(grepl('.mu', sel))){
sel_mu <- gsub(".mu","\\1",sel[grep('mu',sel)])
sel_mu <- gsub("x","\\1",sel_mu)
form_mu <- as.formula(paste("y~",paste(paste0("x.",sel_mu),collapse = '+'), sep= ''))
}else{
sel_mu = 0
form_mu <- as.formula(paste("y~1"))
}
if(any(grepl('sigma', sel))){
sel_sigma <- gsub(".sigma","\\1",sel[grep('sigma', sel)])
sel_sigma <- gsub("x","\\1",sel_sigma)
form_sigma <- as.formula(paste("y~",paste(paste0("x.",sel_sigma),collapse = '+'), sep = ''))
}else{
sel_sigma = 0
form_sigma <- as.formula(paste("y~1"))
}
form <- list(mu = form_mu,
sigma = form_sigma)
glm1 <- glmboostLSS(form, data = ribo[- id,],
method = "noncyclic",
families = GaussianLSS(),
control = boost_control(mstop = 1))
cvr <- cvrisk(glm1, grid = 1:100, papply = lapply)
mstop(glm1) <- mstop(cvr)
coefs <- coef(glm1)
selmu <- length(coef(glm1)$mu)-1
selsi <- length(coef(glm1)$sigma)-1
pred_mu <- predict(glm1$mu, newdata = ribo[id,],
type = "response")
pred_sigma <- predict(glm1$sigma, newdata = ribo[id,],
type = "response")
msep_prob <- (dat[id,'y'] - pred_mu)^2
mae_prob <- abs(dat[id,'y'] - pred_mu)
ls_stab <- dnorm(x = ribo[id,'y'], mean = pred_mu,
sd = pred_sigma, log = TRUE)
return(list(selmu = selmu, selsi = selsi,
mstop = mstop(glm1),
pred_mu = pred_mu, pred_sigma= pred_sigma,
msep = msep_prob, mae = mae_prob,
ls_stab = ls_stab))
}ERG <- mclapply(id, FUN = AMP_LSS_stabsel, mc.cores = 11,
mc.preschedule = FALSE)mean(unlist(sapply(ERG, function(x)x$selmu)))[1] 1.929577mean(unlist(sapply(ERG, function(x)x$selsi)))[1] 0mean(unlist(sapply(ERG, function(x)x$msep)))[1] 0.4384487median(sqrt(unlist(sapply(ERG, function(x)x$msep))))[1] 0.3580849mean(unlist(sapply(ERG, function(x)x$ls_stab)))[1] -1.065873Deselection
AMP_LSS <- function(id)
{
set.seed(id)
print(id)
glm1 <- glmboostLSS( y ~ x, data = dat[- id,],
method = "noncyclic",
families = GaussianLSS(),
control = boost_control(mstop = 1))
cvr <- cvrisk(glm1, grid = 1:100, papply = lapply)
mstop(glm1) <- mstop(cvr)
pred <- predict(glm1, newdata = dat[id,],
type = "response")
msep <- (dat[id,]$y - pred$mu)^2
mae <- abs(dat[id,]$y - pred$mu)
coefs <- coef(glm1, which = "")
selmu <- length(coef(glm1)$mu)-1
selsi <- length(coef(glm1)$sigma)-1
desel <- DeselectBoostLSS_2(glm1, tau = 0.01,
fam = GaussianLSS())
pred_mu <- predict(desel$mu, newdata = ribo[id,],
type = "response")
pred_sigma <- predict(desel$sigma,
newdata = ribo[id,],
type = "response")
msep_desel <- (ribo[id,'y'] - pred_mu)^2
mae_desel <- abs(ribo[id,'y'] - pred_mu)
selmu_desel <- length(coef(desel$mu)) -1
selsi_desel <- length(coef(desel$sigma)) -1
ls_desel <- dnorm(x = ribo[id,'y'],
mean = pred_mu, sd = pred_sigma,
log = TRUE)
return(list(pred = pred, msep = msep,
mstop = mstop(glm1),
selmu = selmu, selsi = selsi,
selmu_desel = selmu_desel,
selsi_desel = selsi_desel,
pred_mu= pred_mu, pred_sigma =pred_sigma,
msep_desel = msep_desel,mae_desel = mae_desel,
ls_desel = ls_desel))
}Which calls the function DeselectBoostLSS_2:
DeselectBoostLSS_2 <-
function(object,
data = NULL,
fam,
tau = NULL,
method = c('attributable', 'cumulative')) {
data = attr(object, 'data')
mstop <-
ifelse(inherits(object, 'nc_mboostLSS'),
mstop(object)[1],
sum(mstop(object)))
if (length(risk(object, merge = T)) > (mstop + 2))
stop("risk cannot contain more entries than mstop")
parameter = names(object)
which.response <- colnames(object[[1]]$response) == colnames(data)
name.response <- colnames(data)[which.response]
select <- selected(object, parameter = names(object))
select1 <- selected(object, parameter = names(object))
if (inherits(object, 'glmboostLSS')) {
select[[1]] <- select[[1]] - 1
select[[2]] <- select[[2]] - 1
select[[1]] <- select[[1]][select[[1]] != 0]
select[[2]] <- select[[2]][select[[2]] != 0]
RiskRed <- risk(object, merge = T)
totalRiskRed <-
(risk(object, merge = T)[1] - risk(object, merge = T)[length(risk(object, merge =
T))])
diffRiskRed_all <-
sapply(2:(length(RiskRed) - 1), function(k) {
RiskRed[k] - RiskRed[k + 1]
})
names(diffRiskRed_all) <- names(risk(object, merge = T)[-c(1:2)])
diffRiskRed <- vector("list")
diffRiskRed[[1]] <-
diffRiskRed_all[names(diffRiskRed_all) == parameter[[1]]][select1[[1]] -
1 != 0]
diffRiskRed[[2]] <-
diffRiskRed_all[names(diffRiskRed_all) == parameter[[2]]][select1[[2]] -
1 != 0]
} else{
RiskRed <- risk(object, merge = T)
diffRiskRed_all <-
sapply(1:(length(RiskRed) - 1), function(k) {
RiskRed[k] - RiskRed[k + 1]
})
diffRiskRed_all <- diffRiskRed_all[-c(1:4)]
names(diffRiskRed_all) <- names(risk(object, merge = T)[-c(1:2)])
diffRiskRed <- vector("list")
diffRiskRed[[1]] <-
diffRiskRed_all[names(diffRiskRed_all) == parameter[[1]]]
diffRiskRed[[2]] <-
diffRiskRed_all[names(diffRiskRed_all) == parameter[[2]]]
}
if (is.null(coef(object)[[1]]))
select[[1]] = 0
else
select[[1]] = select[[1]]
if (is.null(coef(object)[[2]]))
select[[2]] = 0
else
select[[2]] = select[[2]]
Var = lapply(1:length(parameter), function(i) {
count(select[[i]])[2]
})
# If a parameter is not selected
# zero <- sapply(1:length(parameter),function(i){length(select[[i]])})
# Risk.Var <- lapply(which(zero != 0),function(i){sapply(1:dim(Var[[i]])[1],function(j){sum(diffRiskRed[[i]][which(count(select[[i]])[[1]][j] == select[[i]])])})})
Risk.Var <-
lapply(1:2, function(i) {
sapply(1:dim(Var[[i]])[1], function(j) {
sum(diffRiskRed[[i]][which(count(select[[i]])[[1]][j] == select[[i]])])
})
})
# diffRiskRed[[i]]
# which(count(select[[i]])[[1]][j] == select[[i]])
w.parameter <-
c(rep(parameter[1], length(count(select[[1]])[[1]])), rep(parameter[2], length(count(select[[2]])[[1]])))
n.parameter <-
c(names(object[[1]]$coef()), names(object[[2]]$coef()))
if ('(Intercept)' %in% n.parameter)
n.parameter <- n.parameter[-which(n.parameter == '(Intercept)')]
Risk.Var_1 <- unlist(Risk.Var)[!is.na(unlist(Risk.Var))]
if (any(Risk.Var_1 == 0))
Risk.Var_1 <- Risk.Var_1[!Risk.Var_1 == 0]
Risk.order <- data.frame(w.parameter, n.parameter, Risk.Var_1)
colnames(Risk.order) <- c('parameter', 'VarName', 'Risk')
Risk.order <- Risk.order[order(Risk.order$Risk), ]
Risk.order$CumRisk <- cumsum(Risk.order$Risk)
perc <- ifelse(is.null(tau), 0.01, tau)
percRiskRed <- sum(Risk.order$Risk) * perc
if (method[1] == 'attributable') {
RiskRedOver <- Risk.order[which(Risk.order$Risk > percRiskRed), ]
} else{
RiskRedOver <- Risk.order[which(Risk.order$CumRisk > percRiskRed), ]
}
if (is.na(parameter[2]))
parameter[2] <- 0
if (is.na(parameter[1]))
parameter[1] <- 0
# if(length(name.response) > 1) name.response <- paste("cbind(",paste(name.response, collapse = ","),")")
name.response <- "y"
if (any(RiskRedOver$parameter == parameter[1])) {
# if(length(name.response) >1) name.response <- paste("cbind(",paste(name.response, collapse = ","),")")
form1 <-
as.formula(paste(name.response, " ~ ", paste(
gsub("x", "x.", RiskRedOver$VarName[RiskRedOver$parameter == parameter[1]]), collapse = "+"
)))
} else{
form1 <- as.formula(paste(name.response, '~1'))
}
if (any(RiskRedOver$parameter == parameter[2])) {
form2 <-
as.formula(paste(name.response, " ~ ", paste(
gsub("x", "x.", RiskRedOver$VarName[RiskRedOver$parameter == parameter[2]]), collapse = "+"
)))
} else{
form2 <- as.formula(paste(name.response, '~1'))
}
formula <- list(form1, form2)
names(formula) <- names(object)
dfbase <-
environment(environment(environment(object[[1]][["fitted"]])[["RET"]][["baselearner"]][[1]][["model.frame"]])[["ret"]][["model.frame"]])[["df"]]
colnames(data$x) <- paste0("x.", colnames(data$x))
data <- data.frame(cbind(y = data$y, x = data$x))
if (inherits(object, "nc_mboostLSS")) {
if (inherits(object, "glmboostLSS")) {
model_after = glmboostLSS(
formula,
data = data,
families = fam,
method = 'noncyclic',
weights = model.weights(object),
control = boost_control(
mstop = mstop(object),
nu = list(
as.numeric(object[[1]]$control$nu),
as.numeric(object[[2]]$control$nu)
)
)
)
} else{
model_after = gamboostLSS(
formula,
data = data,
families = fam,
method = 'noncyclic',
weights = model.weights(object),
control = boost_control(
mstop = mstop(object),
nu = list(
as.numeric(object[[1]]$control$nu),
as.numeric(object[[2]]$control$nu)
)
)
)
}
} else{
if (inherits(object, "glmboostLSS")) {
model_after = glmboostLSS(
formula,
data = data,
families = fam,
method = 'cyclic',
weights = model.weights(object),
control = boost_control(
mstop = mstop(object),
nu = list(
as.numeric(object[[1]]$control$nu),
as.numeric(object[[2]]$control$nu)
)
)
)
} else{
model_after = gamboostLSS(
formula,
data = data,
families = fam,
method = 'cyclic',
weights = model.weights(object),
control = boost_control(
mstop = mstop(object),
nu = list(
as.numeric(object[[1]]$control$nu),
as.numeric(object[[2]]$control$nu)
)
)
)
}
}
out <- model_after
Coef <- coef(model_after)
deselect_para <-
list(coef = Coef,
tau = perc,
deselectmethod = method[1])
out <- append(x = out, values = deselect_para)
class(out) <- c(class(out))
return(out)
}ERG <- mclapply(id, FUN = AMP_LSS, mc.cores = 11,
mc.preschedule = FALSE)mean(unlist(sapply(ERG, function(x)x$selmu)))[1] 19.14085mean(unlist(sapply(ERG, function(x)x$selsi)))[1] 2.140845mean(unlist(sapply(ERG, function(x)x$msep)))[1] 0.2174377median(sqrt(unlist(sapply(ERG, function(x)x$msep))))[1] 0.2377528median(unlist(sapply(ERG, function(x)x$mstop)))[1] 49mean(unlist(sapply(ERG, function(x)x$ls_desel)))[1] -0.7983609